R offers several ways to reverse a string, include some base R options. We go through a few of those in this post. We’ll also compare the computational time for each method.
Reversing a string can be especially useful in bioinformatics (e.g. finding the reverse compliment of a DNA strand). To get started, let’s generate a random string of 10 million DNA bases (we can do this with the stringi package as well, but for our purposes here, let’s just use base R functions).
set.seed(1)
dna <- paste(sample(c("A", "T", "C", "G"), 10000000, replace = T), collapse = "")1) Base R with strsplit and paste
One way to reverse a string is to use strsplit with paste. This is the slowest method that will be shown, but it does get the job done without needing any packages. In this example, we use strsplit to break the string into a vector of its individual characters. We then reverse this vector using rev. Finally, we concatenate the vector of characters into a string using paste.
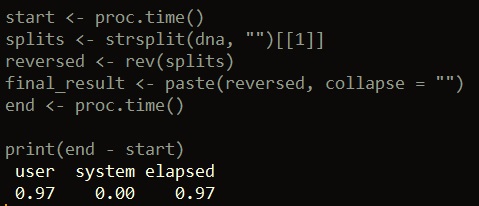
start <- proc.time() splits <- strsplit(dna, "")[[1]] reversed <- rev(splits) final_result <- paste(reversed, collapse = "") end <- proc.time() print(end - start)
2) Base R: Using utf8 magic
This example also does not require any external packages. In this method, we can use the built-in R function utf8ToInt to convert our DNA string to a vector of integers. We then reverse this vector with the rev function. Lastly, we convert this reversed vector of integers back to its original encoding – except now the string is in reverse.
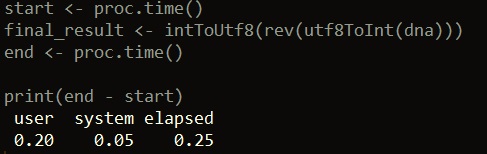
start <- proc.time() final_result <- intToUtf8(rev(utf8ToInt(dna))) end <- proc.time() print(end - start)
3) The stringi package
Of all the examples presented, this option is the fastest when tested. Here we use the stri_reverse function from the stringi package.
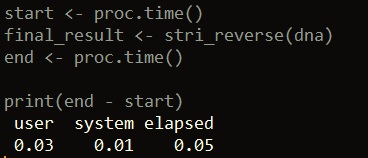
library(stringi) start <- proc.time() final_result <- stri_reverse(dna) end <- proc.time() print(end - start)
4) The Biostrings package
Our last example uses the Biostrings package, which contains a collection of functions useful for working with DNA-string data. One function, called str_rev, can reverse strings. You can download and load the Biostrings package like this:
source("http://bioconductor.org/biocLite.R")
biocLite("Biostrings")
library(Biostrings)Then, all we have to do is input our DNA string into the str_rev function and we get our result.
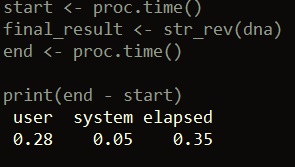
start <- proc.time() final_result <- str_rev(dna) end <- proc.time() print(end - start)
Originally posted on TheAutomatic.net blog.
Disclosure: Interactive Brokers Third Party
Information posted on IBKR Campus that is provided by third-parties does NOT constitute a recommendation that you should contract for the services of that third party. Third-party participants who contribute to IBKR Campus are independent of Interactive Brokers and Interactive Brokers does not make any representations or warranties concerning the services offered, their past or future performance, or the accuracy of the information provided by the third party. Past performance is no guarantee of future results.
This material is from TheAutomatic.net and is being posted with its permission. The views expressed in this material are solely those of the author and/or TheAutomatic.net and Interactive Brokers is not endorsing or recommending any investment or trading discussed in the material. This material is not and should not be construed as an offer to buy or sell any security. It should not be construed as research or investment advice or a recommendation to buy, sell or hold any security or commodity. This material does not and is not intended to take into account the particular financial conditions, investment objectives or requirements of individual customers. Before acting on this material, you should consider whether it is suitable for your particular circumstances and, as necessary, seek professional advice.













Join The Conversation
For specific platform feedback and suggestions, please submit it directly to our team using these instructions.
If you have an account-specific question or concern, please reach out to Client Services.
We encourage you to look through our FAQs before posting. Your question may already be covered!